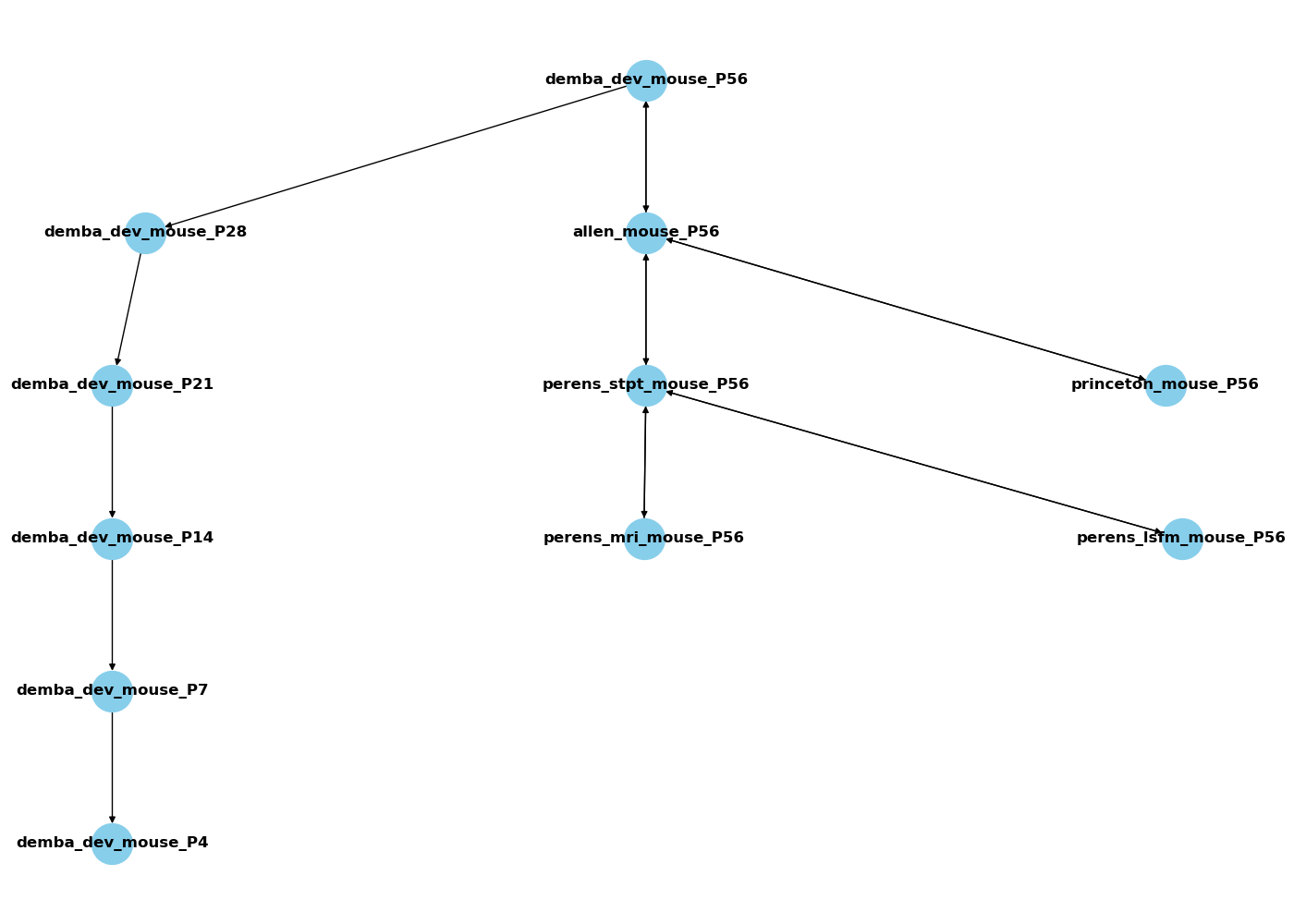
A tool for translating between common coordinate frameworks using deformation matrices. A longstanding problem in NeuroInformatics has been the inability to easily translate data between common coordinate frameworks. CCF_translator aims to solve this. By connecting each new space to an existing one, we can construct a graph of deformations. This means that data can be translated as long as there is a route from one space to another, even if that route passes through multiple other spaces. Now, when new templates for new modalities, strains, or ages are released, users will not be subdivided into unrelated spaces. As long as they are connected to a space which exists in our network, they will be fully connected to all other spaces.
CCF_translator can also interpolate between spaces and create a new intermediate space. This is primarily useful for development, where, for instance, the midpoint between day 5 and day 7 can be taken and used as a postnatal day 6 reference. It could also be useful for making references of disease progression.
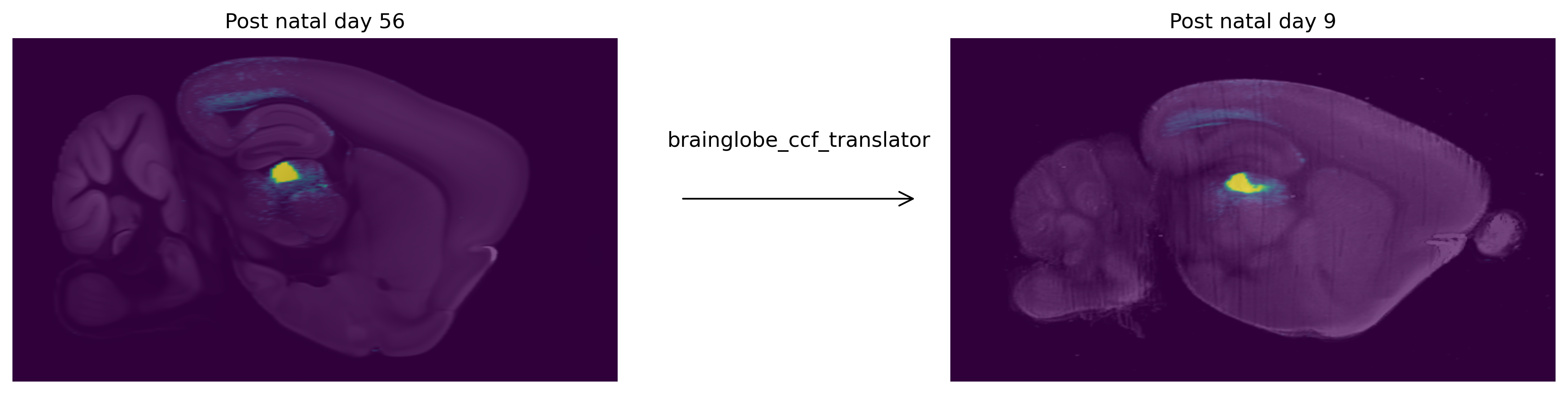
One way you can use CCF_translator is to view data from one space, in another space. For instance the allen connectivity dataset shows projections from viral tracing studies in the adult brain. We can take any of these projection datasets and view them in the developing brain, for instance post natal day 9.
CCF_translator can be installed by running
pip install CCF-translator
the name in CCF_translator aims to copy the name of atlases in the brainglobe_atlasapi when possible.
| Framework Name | name in api | supported age range |
|---|---|---|
| Allen mouse CCFv3 | allen_mouse | 56 |
| Demba developmental mouse | demba_dev_mouse | 4-56 |
| Gubra lightsheet mouse | perens_lsfm_mouse | 56 |
| Gubra MRI mouse | perens_mri_mouse | 56 |
| Gubra STPT mouse | perens_stpt_mouse | 56 |
| Princeton lighsheet mouse | princeton_mouse | 56 |
Transforming points To take a coordinate in one volume and find the equivalent coordinate in a second volume is quite simple in CCF_translator.
import numpy as np
import CCF_translator
points = np.array([(286,250,267), (414,247,452)])
pset = CCF_translator.PointSet(points, 'demba_dev_mouse', voxel_size_micron=20, age_PND=56)
pset.transform(target_age=56, target_space='allen_mouse')
print(f"new points are {pset.values}")new points are [[267 250 286] [452 247 414]]
Transforming volumes
To run the volume examples you will want to install brainglobe-atlasapi using the following
pip install brainglobe-atlasapi
Transforming a volume is equally simple, here we get the volume from the brainglobe api, but you can load it however you like. In the Demba space the valid ages are from 4 to 56, and all of these are valid targets for transformation.
from brainglobe_atlasapi.bg_atlas import BrainGlobeAtlas
import CCF_translator
voxel_size_micron = 10
space_name = r"allen_mouse"
atlas = BrainGlobeAtlas("{space_name}_{voxel_size_micron}um")
source_age = 56
target_age= 32
CCFT_vol = CCF_translator.Volume(
values = atlas.reference,
space = 'allen_mouse',
voxel_size_micron=voxel_size_micron,
segmentation_file=False,
age_PND = source_age
)
CCFT_vol.transform(target_age, 'demba_dev_mouse')
CCFT_vol.save(rf"demo_data/P{target_age}_template_{voxel_size_micron}um.nii.gz")If you would like to add a new space or connect an existing one, please create a deformation matrix and/or describe the required reorientation, flipping, cropping, and padding of the axis between this space and one that already exists in the network, and then open an issue in this repository. Ideally, choose a space which covers all the areas which are covered in your space. While the Allen CCFv3 is very popular, it is missing the anterior olfactory bulb and the caudal portion of the cerebellum and brain stem, so it is not the optimal choice.
CCF_translator was first described in DeMBA: a developmental atlas for navigating the mouse brain in space and time. If you use CCF_translator, please cite that paper.