-
Notifications
You must be signed in to change notification settings - Fork 32
/
optimize.rs
362 lines (334 loc) · 11.6 KB
/
optimize.rs
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
//! To optimize parametric model (non-linear regression)
//!
//! ## `Optimizer` structure
//!
//! ### Declaration
//!
//! ```rust
//! extern crate peroxide;
//! use peroxide::fuga::*;
//! use std::collections::HashMap;
//!
//! pub struct Optimizer<F>
//! where F: Fn(&Vec<f64>, Vec<AD>) -> Option<Vec<AD>> {
//! domain: Vec<f64>,
//! observed: Vec<f64>,
//! func: Box<F>,
//! param: Vec<AD>,
//! max_iter: usize,
//! error: f64,
//! method: OptMethod,
//! option: HashMap<OptOption, bool>,
//! hyperparams: HashMap<String, f64>,
//! }
//! ```
//!
//! ### Method (Should fill)
//!
//! * `new` : Declare new Optimizer. **Should be mutable**
//! * `set_init_param` : Input initial parameter
//! * `set_max_iter` : Set maximum number of iterations
//! * `set_method` : Set method to optimization
//!
//! ### Method (Optional)
//!
//! * `get_domain` : Get domain
//! * `get_error` : Root mean square error
//! * `get_hyperparam` : Get hyperparameter
//! * `set_lr` : Set learning rate (For `GradientDescent`)
//! * `set_lambda_init` : Set initial value of lambda (For `LevenbergMarquardt`)
//! * `set_lambda_max` : Set maximum value of lambda (For `LevenbergMarquardt`)
//!
//! ### Method (Generate result)
//!
//! * `optimize` : Optimize
//!
//! ## Example
//!
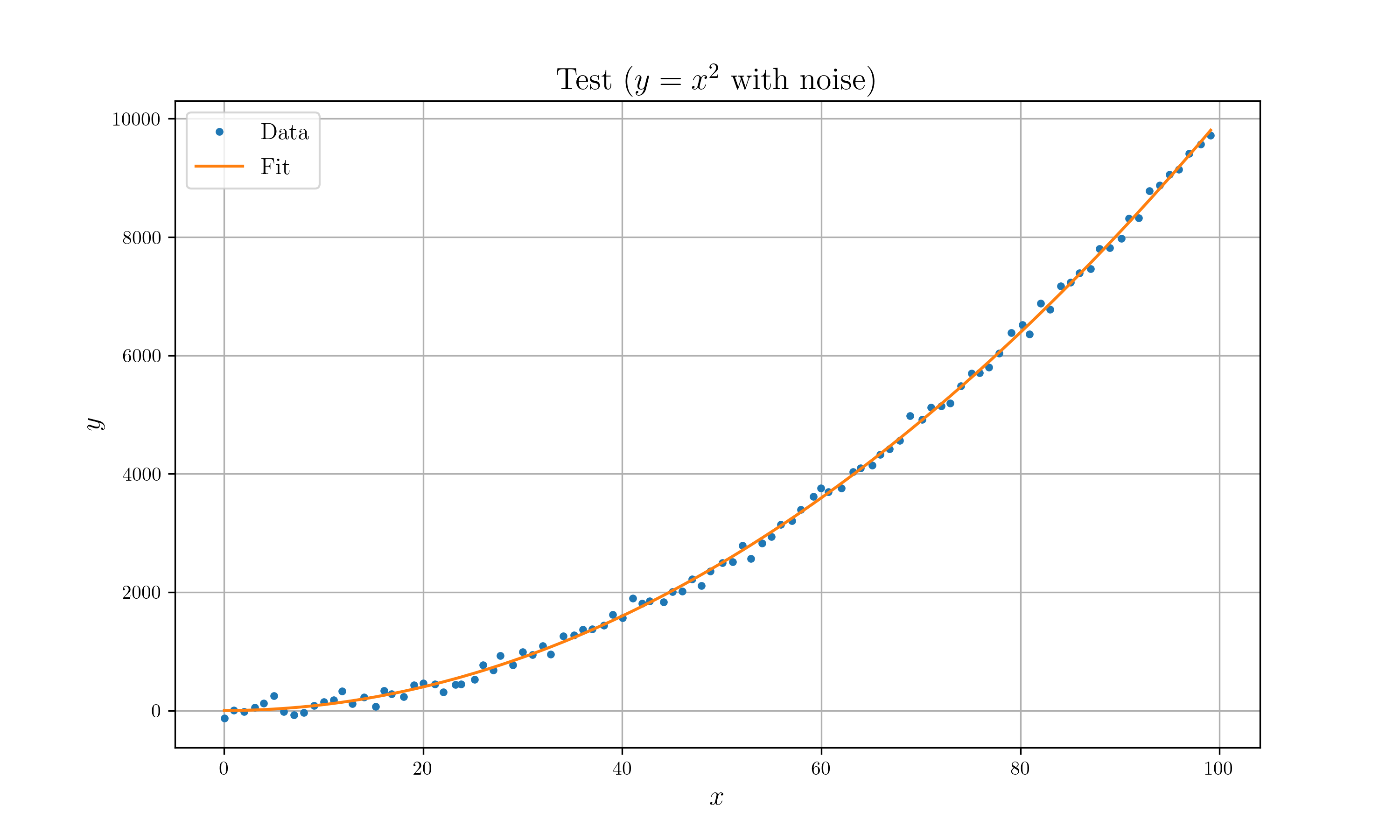
//! * Optimize $y = x^n$ model with $y = x^2$ with gaussian noise.
//!
//! ```rust
//! #[macro_use]
//! extern crate peroxide;
//! use peroxide::fuga::*;
//!
//! fn main() {
//! // To prepare noise
//! let normal = Normal(0f64, 0.1f64);
//! let normal2 = Normal(0f64, 100f64);
//!
//! // Noise to domain
//! let mut x = seq(0., 99., 1f64);
//! x = zip_with(|a, b| (a + b).abs(), &x, &normal.sample(x.len()));
//!
//! // Noise to image
//! let mut y = x.fmap(|t| t.powi(2));
//! y = zip_with(|a, b| a + b, &y, &normal2.sample(y.len()));
//!
//! // Initial parameter
//! let n_init = vec![1f64];
//! let data = hstack!(x.clone(), y.clone());
//!
//! // Optimizer setting
//! let mut opt = Optimizer::new(data, quad);
//! let p = opt.set_init_param(n_init)
//! .set_max_iter(50)
//! .set_method(LevenbergMarquardt)
//! .set_lambda_init(1e-3) // Optional: Set initial value of lambda (Only for `LevenbergMarquardt`)
//! .set_lambda_max(1e+3) // Optional: Set maximum bound of lambda (Only for `LevenbergMarquardt`)
//! .optimize();
//! p.print(); // Optimized parameter
//! opt.get_error().print(); // Optimized RMSE
//!
//! // Plot
//! //#[cfg(feature = "plot")]
//! //{
//! // let z = quad(&x, p.to_ad_vec()).unwrap().to_f64_vec();
//! // let mut plt = Plot2D::new();
//! // plt.set_domain(x)
//! // .insert_image(y) // plot data
//! // .insert_image(z) // plot fit
//! // .set_legend(vec!["Data", "Fit"])
//! // .set_title("Test ($y=x^2$ with noise)")
//! // .set_path("example_data/lm_test.png")
//! // .set_marker(vec![Point, Line])
//! // .savefig().expect("Can't draw a plot");
//! //}
//! }
//!
//! fn quad(x: &Vec<f64>, n: Vec<AD>) -> Option<Vec<AD>> {
//! Some(
//! x.clone().into_iter()
//! .map(|t| AD1(t, 0f64))
//! .map(|t| t.pow(n[0]))
//! .collect()
//! )
//! }
//! ```
//!
//! 
pub use self::OptMethod::{GaussNewton, GradientDescent, LevenbergMarquardt};
use self::OptOption::{InitParam, MaxIter};
use crate::numerical::utils::jacobian;
use crate::structure::ad::{ADVec, AD};
use crate::structure::matrix::Matrix;
use crate::traits::matrix::{LinearAlgebra, MatrixTrait};
use crate::util::useful::max;
use std::collections::HashMap;
#[derive(Debug, Clone, Copy)]
pub enum OptMethod {
GradientDescent,
GaussNewton,
LevenbergMarquardt,
}
#[derive(Debug, Clone, Copy, PartialOrd, PartialEq, Eq, Hash)]
pub enum OptOption {
InitParam,
MaxIter,
}
/// Optimizer for optimization (non-linear regression)
///
/// # Methods
/// * Gradient Descent
/// * Gauss Newton
/// * Levenberg Marquardt
///
/// # Caution
/// * `func` should be boxed. (This allows more generic function)
pub struct Optimizer<F>
where
F: Fn(&Vec<f64>, Vec<AD>) -> Option<Vec<AD>>,
{
domain: Vec<f64>,
observed: Vec<f64>,
func: Box<F>,
param: Vec<AD>,
max_iter: usize,
error: f64,
method: OptMethod,
option: HashMap<OptOption, bool>,
hyperparams: HashMap<String, f64>,
}
impl<F> Optimizer<F>
where
F: Fn(&Vec<f64>, Vec<AD>) -> Option<Vec<AD>>,
{
pub fn new(data: Matrix, func: F) -> Self {
let mut default_option: HashMap<OptOption, bool> = HashMap::new();
default_option.insert(InitParam, false);
default_option.insert(MaxIter, false);
Optimizer {
domain: data.col(0),
observed: data.col(1),
func: Box::new(func),
param: vec![],
max_iter: 0,
error: 0f64,
method: LevenbergMarquardt,
option: default_option,
hyperparams: HashMap::new(),
}
}
/// Get domain
pub fn get_domain(&self) -> Vec<f64> {
self.domain.clone()
}
/// Get error
pub fn get_error(&self) -> f64 {
self.error
}
/// Get hyperparameter (learning rate or lambda or etc.)
pub fn get_hyperparam(&self, key: &str) -> Option<&f64> {
self.hyperparams.get(key)
}
/// Set initial parameter
pub fn set_init_param(&mut self, p: Vec<f64>) -> &mut Self {
if let Some(x) = self.option.get_mut(&InitParam) {
*x = true
}
self.param = p.to_ad_vec();
self
}
/// Set maximum iteration
pub fn set_max_iter(&mut self, n: usize) -> &mut Self {
if let Some(x) = self.option.get_mut(&MaxIter) {
*x = true
}
self.max_iter = n;
self
}
/// Set optimization method
pub fn set_method(&mut self, method: OptMethod) -> &mut Self {
self.method = method;
self
}
/// Set learning rate for `GradientDescent`
pub fn set_lr(&mut self, lr: f64) -> &mut Self {
self.hyperparams.insert("lr".to_string(), lr);
self
}
/// Set initial lambda for `LevenbergMarquardt`
pub fn set_lambda_init(&mut self, lambda_init: f64) -> &mut Self {
self.hyperparams
.insert("lambda_init".to_string(), lambda_init);
self
}
/// Set maximum lambda for `LevenbergMarquardt`
pub fn set_lambda_max(&mut self, lambda_max: f64) -> &mut Self {
self.hyperparams
.insert("lambda_max".to_string(), lambda_max);
self
}
/// Main function for optimization
pub fn optimize(&mut self) -> Vec<f64> {
// Receive initial data
let (x_vec, y_vec) = (self.domain.clone(), self.observed.clone());
let (p_init, max_iter) = (self.param.clone(), self.max_iter);
let safe_f = |p: &Vec<AD>| (self.func)(&x_vec, p.clone()).unwrap();
let unsafe_f = |p: Vec<AD>| (self.func)(&x_vec, p);
// Take various form of initial data
let p_init_vec = p_init.to_f64_vec();
let y = y_vec.into();
// Declare mutable values
let mut p: Matrix = p_init_vec.clone().into();
let mut j = jacobian(safe_f, &p_init_vec);
let mut y_hat: Matrix = safe_f(&p_init).to_f64_vec().into();
let mut jtj = &j.t() * &j;
let mut valid_p = p.clone();
let mut err_stack = 0usize;
match self.method {
GradientDescent => {
let alpha = *self.hyperparams.get("lr").unwrap_or(&1e-3);
for i in 0..max_iter {
let h = alpha * j.t() * (&y - &y_hat);
let p_cand = &p + &h;
match unsafe_f(p_cand.data.to_ad_vec()) {
Some(value) => {
p = p_cand;
valid_p = p.clone();
err_stack = 0;
j = jacobian(safe_f, &p.data);
y_hat = value.to_f64_vec().into();
}
None => {
if i < max_iter - 1 && err_stack < 3 {
p = p_cand;
err_stack += 1;
} else {
p = valid_p;
break;
}
}
}
}
}
GaussNewton => unimplemented!(),
LevenbergMarquardt => {
let mut chi2 = ((&y - &y_hat).t() * (&y - &y_hat))[(0, 0)];
let mut nu = 2f64;
let lambda_0 = *self.hyperparams.get("lambda_init").unwrap_or(&1e-3);
let lambda_max = *self
.hyperparams
.get("lambda_max")
.unwrap_or(&f64::MAX.sqrt());
let mut lambda = lambda_0 * max(jtj.diag());
for i in 0..max_iter {
if lambda > lambda_max {
println!(
"Caution: At {}-th iter, lambda exceeds max value: {}",
i + 1,
lambda
);
break;
}
let h: Matrix;
let b_lu = (jtj.clone() + lambda * jtj.to_diag()).lu();
if b_lu.det() == 0f64 {
break;
}
let b = b_lu.inv();
h = b * j.t() * (&y - &y_hat);
let p_temp = &p + &h;
match unsafe_f(p_temp.data.to_ad_vec()) {
Some(value) => {
let j_temp = jacobian(safe_f, &p.data);
let y_hat_temp: Matrix = value.to_f64_vec().into();
let chi2_temp = ((&y - &y_hat_temp).t() * (&y - &y_hat_temp))[(0, 0)];
let rho = (chi2 - chi2_temp)
/ (h.t()
* (lambda * jtj.to_diag() * h.clone() + j.t() * (&y - &y_hat)))
[(0, 0)];
if rho > 0f64 {
p = p_temp;
valid_p = p.clone();
err_stack = 0;
j = j_temp;
jtj = &j.t() * &j;
y_hat = y_hat_temp;
chi2 = chi2_temp;
lambda *=
max(vec![1f64 / 3f64, 1f64 - (2f64 * rho - 1f64).powi(3)]);
nu = 2f64;
} else {
lambda *= nu;
nu *= 2f64;
}
}
None => {
if i < max_iter - 1 && err_stack < 3 {
p = p_temp;
err_stack += 1;
} else {
p = valid_p;
break;
}
}
}
}
}
}
let error_temp = &y - &y_hat;
self.error = ((error_temp.t() * error_temp)[(0, 0)] / y.row as f64).sqrt();
p.data
}
}